SA14-14-2Rep: A Replicon Vector Derived from the Live-Attenuated Japanese Encephalitis Vaccine SA14-14-2 Virus for Human and Veterinary Medicine
Sang-Im Yun, Byung-Hak Song,Young-Min Lee*
DOI10.21767/2573-0282.100004
Sang-Im Yun, Byung-Hak Song and Young-Min Lee*
Department of Animal, Dairy, and Veterinary Sciences; Utah Science Technology and Research, College of Agriculture and Applied Sciences, Utah State University, Logan, Utah, USA
- *Corresponding Author:
- Young-Min Lee
9830 Old Main Hill, Logan, Utah 84322-9830, USA
Tel: +1-435-797-9667
E-mail: youngmin.lee@usu.edu
Received date: January 14, 2016; Accepted date: February 25, 2016; Published date: February 29, 2016
Citation: Lee YM, Song BH, Yun SI. SA14-14-2Rep: A Replicon Vector Derived from the Live-Attenuated Japanese Encephalitis Vaccine SA14-14-2 Virus for Human and Veterinary Medicine. J Pediatric Infect Dis. 2016, 1:4. doi: 10.21767/2573-0282.100004
Abstract
Japanese encephalitis (JE), an acute inflammatory disease of the brain, is caused by infection with Japanese encephalitis virus (JEV). Children are most susceptible to JEV, which is commonly found in Asia. JEV is an enveloped RNA virus that is genetically similar to West Nile, dengue, and yellow fever viruses. No specific antiviral drugs are currently available to treat JE disease, but four types of vaccine are licensed locally to prevent JEV infection. However, the disease continues to be an important global health priority. Of the four vaccines, the most widely used in JE-endemic countries is the live-attenuated vaccine SA14-14-2. In our previous work, we cloned a genomelength cDNA of SA14-14-2 and rescued recombinant viruses entirely from the cloned cDNA (pBac/SA14-14-2). In the present study, we engineered this functional SA14-14-2 cDNA to construct a self-replicating subgenomic replicon (pBac/SA14-14-2Rep) by deleting the coding region for its two viral envelope glycoproteins, prM and E. After transfection of BHK-21 cells with the replicon RNA transcribed in vitro from pBac/SA14-14-2Rep, a combination of confocal microscopic imaging and immunoblotting showed the transient replication of the replicon RNA in the cytoplasm of RNA-transfected cells. Moreover, transfection of the SA14-14-2 replicon RNA into a BHK-21 cell line stably expressing all three JEV structural proteins (C, prM and E) led to the production of singleround infectious particles that packaged the replicon RNA, with a titter of ~105 infectious units per ml. Our SA14-14-2-based replicon represents a valuable tool for studying JEV RNA replication in low-level bio containment facilities and provides a useful platform for developing new vaccine vectors against an array of human and animal pathogens.
Keywords
Flavivirus; Japanese encephalitis virus; SA14-14-2; Live-attenuated vaccine; Vaccine vector; Replicon
Introduction
Japanese encephalitis virus (JEV) belongs to the genus Flavivirus in the family Flaviviridae [1]. Within the genus, JEV is genetically related to other clinically important human pathogens, such as West Nile (WNV), dengue (DENV), and yellow fever (YFV) viruses [2]. Like all Flaviviruses, JEV is an enveloped RNA virus that has an inner core composed of a plus-strand RNA genome and multiple copies of the capsid (C) protein; this core is surrounded by an outer lipid bilayer with 180 copies of the viral membrane (M) and envelope (E) proteins embedded in it [3,4]. The linear genomic RNA is approximately 11,000 nucleotides long [5] and contains a single long open reading frame (ORF) flanked by two short non-coding regions (NCRs) at the 5’ and 3’ ends that possess cis-acting RNA elements required for viral replication [6,7]. Upon entry into a susceptible host cell, the viral genomic RNA serves as an mRNA for the translation of a polyprotein precursor. This precursor is then co- or post-translationally processed into at least 10 smaller proteins, arranged as NH2-CprM- E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH (prM is the precursor of M, and NS stands for nonstructural protein) [8]. The three N-terminal structural proteins (C, prM and E) are required for the formation of infectious virus particles, and the seven C-terminal nonstructural proteins (NS1 to NS5) are involved in the assembly of viral replication complexes in which viral RNA synthesis takes place [9,10]. Recently, a Cterminally extended form of NS1 (NS1’) has been shown to be expressed through a -1 ribosomal frameshift at the beginning of NS2A [8,11,12]. Viral RNA replication occurs entirely in the cytoplasm of infected cells.
JEV is a zoonotic arbovirus that is maintained in a natural transmission cycle involving a number of vertebrate hosts and mosquito vectors. From the perspective of medical importance, wading birds are the main reservoirs for viral maintenance, domestic pigs are the primary hosts for viral amplification, and culicine mosquitoes (i.e., Culex tritaeniorhynchus) are the principal vectors for viral transmission [13]. Humans become infected through the bite of a JEV-infected mosquito; only a minority of these bites result in clinical disease, ranging from a nonspecific febrile illness to the full-blown Japanese encephalitis (JE), a potentially fatal inflammatory disease of the central nervous system [14,15]. JE occurs throughout most of Asia and parts of the Western Pacific region, with a high likelihood of the disease spreading around the world [16,17]. In endemic areas, JE has a fatality rate of 20-30%, and up to 50% of survivors have permanent neurological and/or psychiatric complications [9]. Currently, there is no clinically approved treatment available for JE, but there are four types of JE vaccine licensed for local use in different countries [9]. Of these, the liveattenuated SA14-14-2 vaccine has become the most widely administered in JE-affected areas. In our previous studies, we determined the complete genome sequence of SA14-14-2, cloned its full-length cDNA into a bacterial artificial chromosome (BAC), and rescued infectious viruses from the cloned cDNA [18,19].
We now report that using our full-length infectious SA14-14-2 cDNA molecular clone, we have succeeded in constructing a transiently self-replicating subgenomic replicon that lacks the viral prM and E genes. We have also demonstrated that transfection of the in vitro synthesized SA14-14-2 replicon RNA into a packaging cell line leads to the production of single-round infectious particles that encapsidate the replicon RNA. Our SA14-14-2-based replicon system offers a new platform for developing a novel viral vector that should be applicable to the development of vaccines against many pathogens of humans and animals.
Materials and Method
Cells
Baby hamster kidney (BHK-21) cells were cultivated in alpha minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1x MEM vitamin solution, 50 units/ml penicillin, and 50 μg/ml streptomycin [20]. Two derivatives of BHK-21 cells (BHK/SinRep19 and BHK/ SinRep19/JEV C-E) were maintained under the same conditions in the presence of 1 μg/ml puromycin [21]. All cell culture reagents were purchased from GIBCO.
Plasmid construction
Using our recently developed full-length infectious cDNA clone of SA14-14-2 (designated pBac/SA14-14-2 [19]), we constructed two subgenomic cDNA clones by overlap extension PCR and standard recombinant DNA techniques. (1) For pBac/SA14-14-2Rep, two overlapping DNA fragments were first amplified by PCR of pBac/SA14-14-2 with two primer pairs, SArep1+SArep2 and SArep3+SArep4, respectively. The two amplified fragments were then combined in a second round of PCR with the outermost primers SArep1 and SArep4, and the 1,066-bp PacI-SphI fragment of the resulting amplicons was ligated with the 2,644-bp SphII-BamHI and 12,863-bp BamHI-PacI fragments of pBac/SA14-14-2. (2) For pBac/SA14-14-2Rep/AAG, the first two DNA fragments were produced by PCR amplification of pBac/SA14-14-2 with two pairs of primers, SArepAAG1+SArepAAG2 and SArepAAG3+SArepAAG4. Again, these two PCR products were joined by a second round of PCR with the outermost primers, SArepAAG1 and SArepAAG4. The 1,395-bp SacII-AatII fragment of the fused amplicons was ligated with the 8,077-bp AatII-PacI and 7,101-bp PacI-SacII fragments of pBac/SA14-14-2Rep. Table 1 provides a list of the primers used in this study.
| Primer | Sequence | Orientation |
|---|---|---|
| SArep1 | 5’- TAGTTAATTAACCTGCAGGGGGCTGTTAGAGG-3’ | Forward |
| SArep2 | 5’- TCAATGGCACATCCAGTGTCGGCTCCTGCACAAGCTATGA- 3’ | Reverse |
| SArep3 | 5’- TCATAGCTTGTGCAGGAGCCGACACTGGATGTGCCATGA- 3’ | Forward |
| SArep4 | 5’- TTTGCATGCTGTTCCAAGCTCTGTGCTCATCA- 3’ | Reverse |
| SArepAAG1 | 5’-GACCCGCGGTTTTGGGAGATGGTTGATGAAGAGA- 3’ | Forward |
| SArepAAG2 | 5’-CAGCGGTTTGACGGCACATCCCGCTGCGCTGATCGCCATCCTGG- 3’ | Reverse |
| SArepAAG3 | 5’-CCAGGATGGCGATCAGCGCAGCGGGATGTGCCGTCAAACCGCTG-3’ | Forward |
| SArepAAG4 | 5’- TCAAGACGTCTTCGTATCTCCTGAGTGAGGTCAT- 3’ | Reverse |
Table 1: List of primers used in this study.
in vitro transcription and RNA transfection
The full-length SA14-14-2 cDNA clone and its two derivatives were digested with XbaI to make linear template DNAs, and then blunted with mung bean nuclease to remove the XbaI-generated 5’ overhangs. The linearized DNAs were purified by phenol-chloroform extraction followed by ethanol precipitation. RNA transcripts were synthesized by in vitro transcription with SP6 RNA polymerase (New England Biolabs) in the presence of the cap analog m7G(5’)ppp(5’)A (New England Biolabs), as described previously [19]. The integrity and yield of the RNA transcripts were determined by agarose gel electrophoresis and the incorporation rate of [3H]UTP, respectively [22]. Under our optimized conditions, RNAs were transfected into BHK-21 or BHK-21-derived cell lines by electroporation using a BTX model ECM 830 electroporator (Harvard Apparatus), as described elsewhere [6,7,20].
Immunofluorescence assay (IFA)
Cells (1 x 105) grown on 4-well glass bottom chamber slides (Lab-Tek) were washed in phosphate-buffered saline (PBS) and fixed/permeabilized with cold methanol at -20°C for 10 min. The cells were washed with PBS (3 x 10 min) and blocked with 5% bovine serum albumin (BSA) in PBS at room temperature for 30 min. After three 10-min washes in PBS, they were incubated for 1 h at room temperature with a primary antibody diluted in PBS containing 2.5% BSA. The primary antibodies used in this indirect IFA were four rabbit polyclonal antisera raised against JEV C (α-C; 1:2,000 dilution), M (α-M; 1:250 dilution), E (α-EN-term; 1:2,000 dilution), and NS1 (α- NS1N-term; 1:2,000 dilution) [8]. After three 10-min washes in PBS, the cells were again incubated for 1 h at room temperature with a secondary Cy3-conjugated goat anti-rabbit antibody (1:1,000 dilution; Molecular Probes) diluted in PBS containing 2.5% BSA. After three 10-min washes in PBS, the cells were counterstained with 300 nM 4’,6-diamidino-2- phenylindole (DAPI, Molecular Probes) in PBS at room temperature for 5 min. They were then washed with PBS (2 x 10 min), mounted using an antifade mounting medium (Dako), and examined using an LSM-710 confocal microscope controlled by the Zen 2010 software (Carl Zeiss).
Immunoblot analysis
Cells (3 x 105) plated in 6-well plates were lysed with 200 μl of 1x sample buffer (80 mM Tri-HCl [pH 6.8], 2.0% SDS, 10% glycerol, 0.1 M DTT, and 0.2% bromophenol blue). One-tenth of each total cell lysate was separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Millipore), and blocked with 5% nonfat dried milk in wash buffer (0.2% Tween 20 in PBS) at room temperature for 1 h. After three 10-min washes in wash buffer, the blots were incubated at room temperature for 2 h with one of the following primary antibodies [8,23]: mouse anti-JEV hyperimmune ascitic fluid (α-JEV; 1:1,000 dilution), rabbit anti- C antiserum (α-C; 1:3,000 dilution), rabbit anti-M antiserum (α-M; 1:1,000 dilution), rabbit anti-E antiserum (α-EN-term; 1:4,000 dilution), rabbit anti-NS1 antiserum (α-NS1N-term; 1:3,000 dilution), or rabbit anti-GAPDH antiserum (α-GAPDH; 1:5,000 dilution). After three 10-min washes in wash buffer, the blots were again incubated at room temperature for 2 h with an alkaline phosphatase (AP)-conjugated secondary antibody against mouse or rabbit IgG (Jackson Immuno Research; 1:5,000 dilution). They were then washed three times and developed by incubating with a mixture of 5- bromo-4-chloro-3-indolyl phosphate (BCIP) and nitro blue tetrazolium (NBT).
Results
Construction of a subgenomic replicon based on the live-attenuated JE vaccine SA14-14-2 virus
Our recent study reported the first development of a genetically stable full-length infectious cDNA clone for SA14-14-2, designated pBac/SA14-14-2 [19]. For this purpose we employed the single-copy BAC vector pBeloBAC11, which was shown to be able to house the large insert of the ~11-kb full-length JEV cDNA in E. coli [20]. In the present work, we aimed to construct a replication-competent, propagationdeficient viral replicon as a new vaccine platform based on our functional SA14-14-2 cDNA. To this end, we introduced an internal in-frame deletion of 2,001 nucleotides into pBac/ SA14-14-2 to remove all of the coding sequence for the two viral surface glycoproteins prM and E, thereby creating pBac/SA14-14-2Rep (Figure 1a). In this replicon, the correct membrane insertion and biogenesis of the viral NS1 protein was ensured by fusing the last 22 amino acid residues of the viral C protein, which act as an internal signal sequence originally for the subsequent protein prM, directly to the Nterminus of NS1. As a negative control, we also generated a replication-defective replicon, pBac/SA14-14-2Rep/AAG (Figure 1a), in which the critical Gly-Asp-Asp (GDD) motif at positions 667 to 669 in the viral NS5 polymerase active site were replaced by Ala-Ala-Gly (AAG).
Figure 1: Construction of a subgenomic replicon of the liveattenuated JE vaccine SA14-14-2 virus. (a) Schematic diagram of the SA14-14-2 replicon vectors used in this study. The genome-length infectious cDNA clone of SA14-14-2 (pBac/SA14-14-2) was used to generate two subgenomic cDNA constructs, pBac/SA14-14-2Rep and pBac/ SA14-14-2Rep/AAG. The viral ORF is shown, along with black horizontal bars at both termini that represent the 5’ and 3’ NCRs. White vertical lines represent major cleavage sites within the color-coded polyprotein. An asterisk indicates a site of the GDD-to-AAG mutation introduced into the NS5 polymerase active site. (b) Synthesis of SA14-14-2 replicon RNAs by in vitro run-off transcription. Each of the three linearized cDNAs was transcribed by SP6 RNA polymerase. Aliquots of the transcription reactions were run on a 0.6% agarose gel containing ethidium bromide. The 3-kb DNA band serves as a reference band.
We used each of the three recombinant cDNAs (pBac/ SA14-14-2, pBac/SA14-14-2Rep, and pBac/SA14-14-2Rep/AAG) as a template for in vitro run-off ranscription with SP6 RNA polymerase under our optimized experimental conditions [19,20], after linearization by Xba I digestion and subsequent mung bean nuclease treatment to remove the 5’ overhang CTAG left by the Xba I digestion. Following transcription, the integrity of the RNA transcripts was examined by agarose gel electrophoresis. On a 0.6% agarose gel, the genome-length RNA transcribed from pBac/SA14-14-2 migrated just below the 3-kb marker DNA band (Figure 1b), as we described previously [22]. We noted that the subgenomic RNA made from pBac/ SA14-14-2Rep ran slightly faster in the gel (Figure 1b) than did the genome-length RNA derived from pBac/SA14-14-2 (used as a reference), in agreement with its smaller size resulting from the deletion of the viral prM and E genes. As expected, the RNA derived from pBac/SA14-14-2Rep/AAG co-migrated precisely with that synthesized from pBac/SA14-14-2Rep.
Characterization of the subgenomic replicon SA14-14-2Rep for transient replication and gene expression in mammalian cells
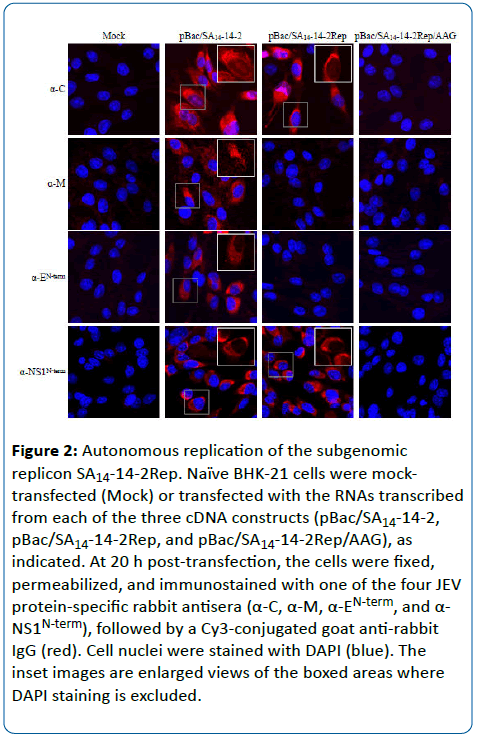
To examine the ability of the subgenomic SA14-14-2Rep or SA14-14-2Rep/AAG replicon to establish autonomous replication in cells, the corresponding in vitro-synthesized RNAs were transfected into naïve BHK-21 cells by electroporation, and viral protein expression was then monitored by IFA with four rabbit polyclonal antisera specific for the JEV C (α-C), M (α-M), E (α-EN-term), and NS1 (α-NS1Nterm) proteins. As a positive control, the full-length RNA derived from pBac/SA14-14-2 was included in parallel. At 20 h post-transfection, we observed clear expression of the C and NS1 proteins, but not of prM and E, in cells transfected with the SA14-14-2Rep RNA, predominantly localized to the cytoplasmic and perinuclear compartments (Figure 2). The positive and negative controls behaved as expected, in that all four viral proteins tested were readily detected in the SA14-14-2 RNA-transfected cells, and none was detectable in the SA14-14-2Rep/AAG RNA-transfected cells (Figure 2). Interestingly, it was noted that unlike the full-length SA14-14-2 RNA, the subgenomic SA14-14-2Rep RNA did not induce apparent cytopathic effects even at 72 h post-transfection, although both RNAs were replication-competent (data not shown).
Figure 2: Autonomous replication of the subgenomic replicon SA14-14-2Rep. Naïve BHK-21 cells were mocktransfected (Mock) or transfected with the RNAs transcribed from each of the three cDNA constructs (pBac SA14-14-2, pBac SA14-14-2Rep, and pBac SA14-14-2Rep/AAG), as indicated. At 20 h post-transfection, the cells were fixed, permeabilized, and immunostained with one of the four JEV protein-specific rabbit antisera (α-C, α-M, α-EN-term, and α- NS1N-term), followed by a Cy3-conjugated goat anti-rabbit IgG (red). Cell nuclei were stained with DAPI (blue). The inset images are enlarged views of the boxed areas where DAPI staining is excluded.
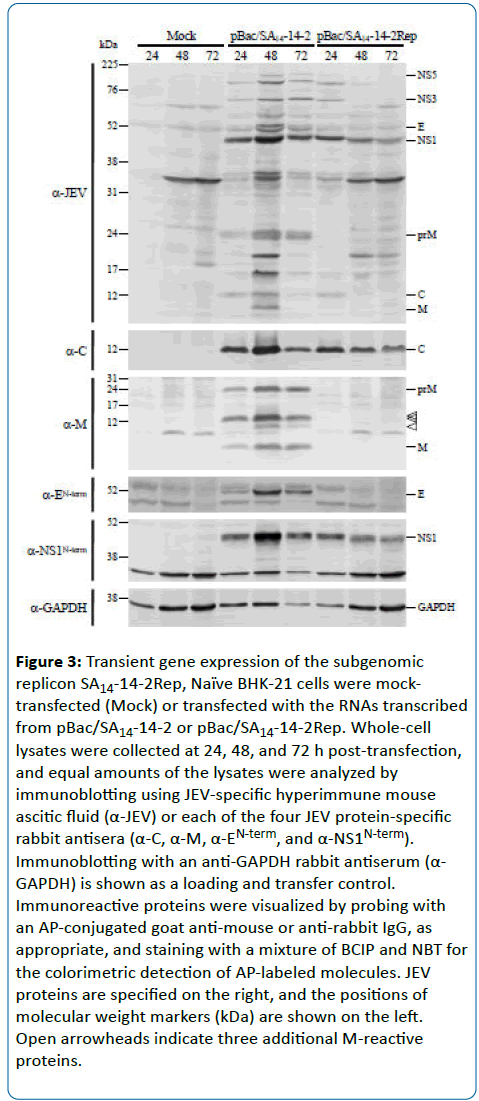
To evaluate the proteolytic processing at the junction between the C and NS1 regions of the polyprotein precursor produced from the subgenomic replicon SA14-14-2Rep, total cell lysates of BHK-21 cells mock-transfected or transfected with the replicon SA14-14-2Rep (or the full-length SA14-14-2 RNA as a positive control) were harvested at 24, 48, and 72 h post-transfection and subjected to immunoblotting with the hyperimmune mouse ascitic fluid raised against JEV and the same four rabbit antisera specifically recognizing the JEV C, M, E, and NS1 proteins. At 24 h post-transfection, both the ~12- kDa C protein and the ~45-kDa NS1 protein were clearly detected in the SA14-14-2Rep RNA-transfected cells, as seen in the SA14-14-2 RNA-transfected cells (Figure 3). However, the accumulation levels of these proteins in the SA14-14-2Rep RNA-transfected cells gradually decreased over the 72-h period of study because of the lack of viral spread, while their levels in the SA14-14-2 RNA-transfected cells were increased at 48 h post-transfection as a result of viral spread, but dramatically decreased at 72 h post-transfection as a result of virus-induced cell death (Figure 3). Also, we confirmed that there was no expression of the prM or E proteins in the SA14-14-2Rep RNA-transfected cells. Our results indicate that SA14-14-2Rep represents a functional JEV replicon that is capable of initiating RNA replication and gene expression but lacks the ability to produce infectious virus.
Figure 3: Transient gene expression of the subgenomic replicon SA14-14-2Rep, Naïve BHK-21 cells were mocktransfected (Mock) or transfected with the RNAs transcribed from pBac SA14-14-2 or pBac SA14-14-2Rep. Whole-cell lysates were collected at 24, 48, and 72 h post-transfection, and equal amounts of the lysates were analyzed by immunoblotting using JEV-specific hyperimmune mouse ascitic fluid (α-JEV) or each of the four JEV protein-specific rabbit antisera (α-C, α-M, α-EN-term, and α-NS1N-term). Immunoblotting with an anti-GAPDH rabbit antiserum (α- GAPDH) is shown as a loading and transfer control. Immunoreactive proteins were visualized by probing with an AP-conjugated goat anti-mouse or anti-rabbit IgG, as appropriate, and staining with a mixture of BCIP and NBT for the colorimetric detection of AP-labeled molecules. JEV proteins are specified on the right, and the positions of molecular weight markers (kDa) are shown on the left. Open arrowheads indicate three additional M-reactive proteins.
Production of propagation-deficient singleround infectious SA14-14-2 particles by transcomplementation
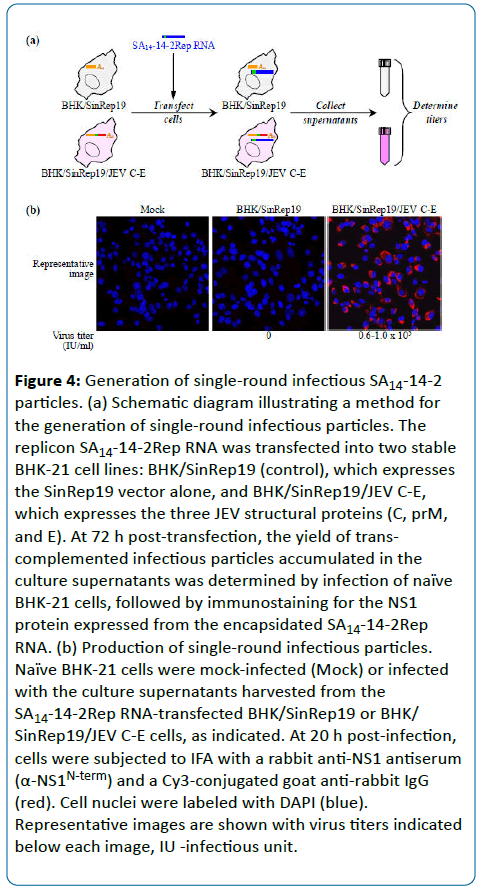
To investigate whether the replication-competent subgenomic SA14-14-2Rep RNA can be packaged into singleround infectious particles when the missing prM and E structural proteins are provided in trans, we took advantage of a stable BHK-21 cell line (BHK/SinRep19/JEV C-E) we had previously developed [21] that constitutively expresses all three JEV structural proteins (C, prM, and E). This cell line persistently carries a noncytopathic Sindbis virus (SINV) replicon RNA that contains two additional cistrons driven by its own subgenomic promoter, one for the three JEV structural proteins and the other for the puromycin resistance gene as a dominant selectable marker. As a control, we used another stable BHK-21 cell line (BHK/SinRep19) harboring the SINV vector replicon RNA without any insertion of JEV genes [21].
We transfected the SA14-14-2Rep RNA into the BHK/ SinRep19/JEV C-E or BHK/SinRep19 cell line. At 72 h posttransfection, the culture supernatants were collected and used to infect naïve BHK-21 cells. We then quantitated the number of cells expressing the NS1 protein from the packaged SA14-14-2Rep RNA by IFA using the rabbit anti-NS1 antiserum (Figure 4a). As presented in Figure 4b, the yield of infectious SA14-14-2 particles from the RNA-transfected BHK/ SinRep19/JEV C-E cells was estimated to be 0.6-1.0 x 105 infectious units/ml. As expected, no infectious particles were detected in the supernatant from the RNA-transfected BHK/ SinRep19 cells. We also confirmed that there was no evidence of RNA recombination that would potentially generate propagation-competent infectious virions, as shown by the reinfection of naïve BHK-21 cells with the culture supernatant harvested from replicon particle-infected BHK-21 cells (data not shown).
Figure 4: Generation of single-round infectious SA14-14-2 particles. (a) Schematic diagram illustrating a method for the generation of single-round infectious particles. The replicon SA14-14-2Rep RNA was transfected into two stable BHK-21 cell lines: BHK/SinRep19 (control), which expresses the SinRep19 vector alone, and BHK/SinRep19/JEV C-E, which expresses the three JEV structural proteins (C, prM, and E). At 72 h post-transfection, the yield of transcomplemented infectious particles accumulated in the culture supernatants was determined by infection of naïve BHK-21 cells, followed by immunostaining for the NS1 protein expressed from the encapsidated SA14-14-2Rep RNA. (b) Production of single-round infectious particles. Naïve BHK-21 cells were mock-infected (Mock) or infected with the culture supernatants harvested from the SA14-14-2Rep RNA-transfected BHK/SinRep19 or BHK/ SinRep19/JEV C-E cells, as indicated. At 20 h post-infection, cells were subjected to IFA with a rabbit anti-NS1 antiserum (α-NS1N-term) and a Cy3-conjugated goat anti-rabbit IgG (red). Cell nuclei were labeled with DAPI (blue). Representative images are shown with virus titers indicated below each image, IU -infectious unit.
Discussion
Previously, we developed the first full-length infectious cDNA clone of SA14-14-2 [19], a live-attenuated JE vaccine that has been widely used for human immunization in several JEendemic countries [9]. In the present study, we have engineered the functional genome-length SA14-14-2 cDNA to create a self-replicating subgenomic replicon SA14-14-2Rep lacking its two viral surface glycoproteins, prM and E. After transfection into permissive BHK-21 cells, we demonstrated the ability of the SA14-14-2Rep RNA to establish transient replication and gene expression, with no concomitant production of infectious particles. Furthermore, we have shown that the replication-competent SA14-14-2Rep RNA is capable of being encapsidated into propagation-deficient single-round infectious particles when the missing prM and E proteins are supplied in trans. Altogether, this SA14-14-2Rep replicon system can help us elucidate the molecular basis of JEV RNA replication without the use of high-level biocontainment facilities, and it provides a powerful new tool for delivering and expressing foreign genes of interest for modern molecular medicine.
Over the last few decades, a number of plus-strand RNA viruses, particularly alphaviruses and Flaviviruses, have been extensively investigated for use as viral vectors, not only for the expression of heterologous genes in a variety of animal cells but also for the development of novel antiviral and anticancer vaccines [24]. The foundation of this RNA-based viral vector approach was laid in the late 1980s, when Rice and co-workers first succeeded in cloning a full-length infectious cDNA of SINV (an alphavirus) [25], creating a replicon by replacing the viral structural protein-coding region with a reporter gene [26], and then making a chimeric virus by introducing a second subgenomic promoter, which drives the expression of a heterologous protein [27,28]. Since then, the same strategy has been applied to other alphaviruses, such as Semliki forest virus and Venezuelan equine encephalitis virus (VEEV). These alphavirus-based vectors have been employed as a vaccine platform and evaluated in preclinical or clinical studies for a broad range of pathogens, including human immunodeficiency virus, human papilloma virus, hepatitis C virus, influenza A virus, Marburg virus, Plasmodium yoelli, and Bacillus anthracis [29]. Moreover, the SINV-based vector has been developed for both short-term transient and long-term stable protein expression [30]. Recently, the VEEV-based replicon has been modified to encode subgenomic RNAs, which are not only transcribed from the subgenomic promoter but also amplified by the previously underutilized viral replicases [31].
Among Flaviviruses, the virus used for the live-attenuated YF vaccine 17D was the first and best studied for use in developing a viral vector as a vaccine platform [32]. The YF17D vaccine is considered to be one of the most effective human vaccines, with an outstanding safety profile, making it an attractive candidate for creating a viral vaccine vector. In 1989, an important milestone was achieved by producing infectious YF17D virus from an in vitro-ligated cDNA template [33]. Since then, the YF17D virus has been used as a backbone to generate several chimeric virus vaccines by replacing its prM and E protein coding region with that of other Flaviviruses, such as JEV, WNV, and four different serotypes of DENV [34]. Of these, the YF-JE chimera is the first viral vector vaccine that has been approved for human use to date in Australia, Malaysia, the Philippines, and Thailand [9]; the other two chimeric vaccines (YF-WN and YF-DEN) are in late-phase clinical studies [34]. Similarly, other Flaviviruses have been used to exchange their prM and E genes or to express and deliver an antigen from unrelated pathogens [29].
RNA virus-based vectors have a broad spectrum of applications, ranging from vaccination to gene therapy for genetic diseases, including cancer. The major advantages of using such a viral vector include: (1) a relatively small genome size, which can be readily manipulated using infectious cDNA technology; (2) cytoplasmic RNA amplification, which prevents nuclear involvement and promotes high levels of gene expression; and (3) synthesis of double-stranded RNA intermediates during genome replication, which enhances antigen-specific immune responses [24,35,36]. However, several limitations have to be overcome in order for the use of the RNA virus-based vectors to be further expanded, particularly with respect to their limited packaging capacity and biosafety-related issues. In this regard, our SA14-14-2- based replicon is a promising platform for developing a new vaccine vector, because SA14-14-2 is capable of packaging recombinant genomes as large as ~15 kb [21] and does not require high-level biocontainment facilities (biosafety levels 3 and 4) [37]. We expect that the full-length infectious SA14-14-2 cDNA we developed previously [19] and its subgenomic replicons we created in the present study will offer new opportunities for basic and clinical research directed toward the development of novel JEV-based vaccine vectors [21,38,39].
Acknowledgments
This study was supported by grants from Utah Science Technology and Research (USTAR A35813) and the Utah Agricultural Experiment Station (UAES UTAO1102). This research was approved as journal paper number UAES #8867. We thank Dr. Deborah McClellan for her careful and critical reading of the manuscript.
References
- Thiel HJ, Collett MS, Gould EA, Heinz FX, Houghton M, Meyers G, Purcell RH, Rice CM (2005) Family Flaviviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: 8th report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press, pp. 981-998.
- Lindenbach BD, Thiel HJ, Rice CM (2007) Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins, pp. 1101-1152.
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH (2002) Structure of dengue virus: implications for Flavivirus organization, maturation, and fusion. Cell 108: 717-725.
- Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ (2003) Structure of West Nile virus. Science 302: 248.
- Yun SI, Kim SY, Choi WY, Nam JH, Ju YR, Park KY, Cho HW, Lee YM (2003) Molecular characterization of the full-length genome of the Japanese encephalitis viral strain K87P39. Virus Res 96: 129-140.
- Yun SI, Choi YJ, Song BH, Lee YM (2009) 3' cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions. J Virol 83: 7909-7930.
- Song BH, Yun SI, Choi YJ, Kim JM, Lee CH, Lee YM (2008) A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for Flavivirus RNA replication. RNA 14: 1791-1813
- Kim JK, Kim JM, Song BH, Yun SI, Yun GN, Byun SJ, Lee YM (2015) Profiling of viral proteins expressed from the genomic RNA of Japanese encephalitis virus using a panel of 15 region-specific polyclonal rabbit antisera: implications for viral gene expression. PLoS One 10: e0124318.
- Yun SI, Lee YM (2014) Japanese encephalitis: the virus and vaccines. Hum VaccinImmunother 10: 263-279.
- Yun SI, Lee YM (2006) Japanese encephalitis virus: molecular biology and vaccine development. In: Kalitzky M, Borowski P, editors. Molecular biology of the Flavivirus. Norwich, UK: Horizon Scientific Press, pp. 225-271.
- Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, Hall R, Khromykh AA (2010) NS1' of Flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84: 1641-1647.
- Firth AE, Atkins JF (2009) A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis Flaviviruses suggests NS1' may derive from ribosomal frameshifting. Virol J 6: 14.
- Gubler DJ, Kuno G, Markoff L (2007) Flaviviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins, pp. 1153-1252
- Solomon T (2006) Control of Japanese encephalitis - within our grasp? N Engl J Med 355: 869-871.
- Solomon T, Vaughn DW (2002) Pathogenesis and clinical features of Japanese encephalitis and West Nile virus infections. Curr Top MicrobiolImmunol 267: 171-194.
- Mackenzie JS, Gubler DJ, Petersen LR (2004) Emerging Flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med 10: S98-109.
- Weaver SC, Barrett AD (2004) Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2: 789-801.
- Song BH, Yun GN, Kim JK, Yun SI, Lee YM (2012) Biological and genetic properties of SA14-14-2, a live-attenuated Japanese encephalitis vaccine that is currently available for humans. J Microbiol 50: 698-706.
- Yun SI, Song BH, Kim JK, Yun GN, Lee EY, Li L, Kuhn RJ, Rossmann MG, Morrey JD, Lee YM (2014) A molecularly cloned, live-attenuated Japanese encephalitis vaccine SA14-14-2 virus: a conserved single amino acid in the ij hairpin of the viral E glycoprotein determines neurovirulence in mice. PLoSPathog 10: e1004290.
- Yun SI, Kim SY, Rice CM, Lee YM (2003) Development and application of a reverse genetics system for Japanese encephalitis virus. J Virol 77: 6450-6465.
- Yun SI, Choi YJ, Yu XF, Song JY, Shin YH, Ju YR, Kim SY, Lee YM (2007) Engineering the Japanese encephalitis virus RNA genome for the expression of foreign genes of various sizes: implications for packaging capacity and RNA replication efficiency. J Neurovirol 13: 522-535.
- Yun SI, Song BH, Kim JK, Lee YM (2015) Bacterial artificial chromosomes: a functional genomics tool for the study of positive-strand RNA viruses. J Vis Exp 106: e53164
- Kim JM, Yun SI, Song BH, Hahn YS, Lee CH, Oh HW, Lee YM (2008) A single N-linked glycosylation site in the Japanese encephalitis virus prM protein is critical for cell type-specific prM protein biogenesis, virus particle release, and pathogenicity in mice. J Virol 82: 7846-7862.
- Khromykh AA (2000) Replicon-based vectors of positive-strand RNA viruses. CurrOpinMolTher 2: 555-569.
- Rice CM, Levis R, Strauss JH, Huang HV (1987) Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol 61: 3809-3819.
- Xiong C, Levis R, Shen P, Schlesinger S, Rice CM, Huang HV (1989) Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243: 1188-1191.
- Raju R, Huang HV (1991) Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J Virol 65: 2501-2510.
- Hahn CS, Hahn YS, Braciale TJ, Rice CM (1992) Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. ProcNatlAcadSci USA 89: 2679-2683.
- Mogler MA, Kamrud KI (2015) RNA-based viral vectors. Expert Rev Vaccines 14: 283-312.
- Agapov EV, Frolov I, Lindenbach BD, Pragai BM, Schlesinger S, Rice CM (1998) NoncytopathicSindbis virus RNA vectors for heterologous gene expression. ProcNatlAcadSci USA 95: 12989-12994.
- Kim DY, Atasheva S, McAuley AJ, Plante JA, Frolova EI, Beasley DW, Frolov I (2014) Enhancement of protein expression by alphavirus replicons by designing self-replicating subgenomic RNAs. ProcNatlAcadSci USA 111: 10708-10713.
- >Bonaldo MC, Sequeira PC, Galler R (2014) The yellow fever 17D virus as a platform for new live attenuated vaccines. Hum VaccinImmunother 10: 1256-1265.
- Rice CM, Grakoui A, Galler R, Chambers TJ (1989) Transcription of infectious yellow fever RNA from full-length cDNAtemplates produced by in vitro ligation. New Biol 1: 285-296.
- Monath TP, Seligman SJ, Robertson JS, Guy B, Hayes EB, Condit RC, Excler JL, Mac LM, Carbery B, Chen RT, Brighton Collaboration Viral Vector Vaccines Safety Working Group (2015) Live virus vaccines based on a yellow fever vaccine backbone: standardized template with key considerations for a risk/benefit assessment. Vaccine 33: 62-72.
- Rollier CS, Reyes-Sandoval A, Cottingham MG, Ewer K, Hill AV (2011) Viral vectors as vaccine platforms: deployment in sight. CurrOpinImmunol 23: 377-382.
- Ulmer JB, Mason PW, Geall A, Mandl CW (2012) RNA-based vaccines. Vaccine 30: 4414-4418.
- Centers for Disease Control and Prevention/National Institutes of Health (2009) Biosafety in microbiological and biomedical laboratories (BMBL) 5th Edition.
- Yun SI, Song BH, Koo Y, Jeon I, Byun SJ, Park JH, Joo YS, Kim SY, Lee YM (2009) Japanese encephalitis virus-based replicon RNAs/particles as an expression system for HIV-1 Pr55Gag that is capable of producing virus-like particles. Virus Res 144: 298-305.
- Li SH, Li XF, Zhao H, Deng YQ, Yu XD, Zhu SY, Jiang T, Ye Q, Qin ED, Qin CF (2013) Development and characterization of the replicon system of Japanese encephalitis live vaccine virus SA14-14-2. Virol J 10: 64.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences