Aetiology of Diarrhoea in Children Under Five in Zambia Detected Using Luminex xTAG Gastrointestinal Pathogen Panel
Caroline Cleopatra Chisenga, Samuel Bosomprah*, Natasha Makabilo Laban, Katayi Mwila- Kazimbaya, John Mwaba, Michelo Simuyandi and Roma Chilengi
DOI10.21767/2573-0282.100064
Caroline Cleopatra Chisenga1, Samuel Bosomprah1,2*, Natasha Makabilo Laban1, Katayi Mwila-Kazimbaya1,3, John Mwaba1,3, Michelo Simuyandi1 and Roma Chilengi1
1Research Division, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia
2Department of Biostatistics, School of Public Health, University of Ghana, Legon, Accra
3Department of Biomedical Sciences, School of Health Sciences, University of Zambia, Lusaka, Zambia
- *Corresponding Author:
- Samuel Bosomprah, PhD
Research Division, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia
Tel: +260968356711
E-mail: sbosomprah@gmail.com
Received date: June 12, 2018; Accepted date: June 20, 2018; Published date: June 27, 2018
Citation: Chisenga CC, Bosomprah S, Laban NM, Mwila-Kazimbaya K, Mwaba J, et al. (2018) Aetiology of Diarrhoea in Children under Five in Zambia Detected Using Luminex xTAG Gastrointestinal Pathogen Panel. Pediatric Infect Dis Vol 3:8. doi: 10.21767/2573-0282.100064
Copyright: © 2018 Chisenga CC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: We aimed to document viral, bacterial, and protozoan enteric pathogens responsible of causing moderate-to-severe diarrhoea among children under five presenting at public health facilities in Zambia following the introduction of rotavirus vaccination.
Methods: This was a cross-sectional study in which clinical data and stool samples were collected between July 2012 and October 2013 from children under five years presenting to outpatient clinics in Lusaka province with moderate-tosevere diarrhoea. The study was conducted during the early months post rotavirus vaccine introduction in Zambia. We used Luminex x-TAG® gastrointestinal pathogen panel to simultaneously detect enteric viruses, bacteria and protozoa from the stool samples. We applied the population attributable fraction to estimate pathogen-specific burden of moderate-to-severe diarrhoea.
Results: We analysed 1,135 unique stool samples with clinical data, of which 56% had received one or full dose rotavirus vaccination. The median age was 14 months (IQR=8, 22). The prevalence of moderate-to-severe diarrhoea was estimated as 18.9% (95%CI=16.7, 21.2). The most attributable cases of moderate-to-severe diarrhoea were due to rotavirus {attributable fraction=24.5%; 95%CI=(5.4, 39.7)} followed by Shigella spp. {attributable fraction=6.7%; 95%CI=(0.1, 15.5)}. The top 5 enteric pathogens detected among children were rotavirus (67.6%), Adenovirus (41.5%), ETEC (40.7%), Salmonella (38.4%) and Giardia (37.0%).
Conclusion: We found that about one-third of moderate-to-severe diarrhoea among children were attributable to rotavirus and shigella spp.
Keywords
Enteric pathogens; Gastrointestinal pathogen panel; Diarrhoea; Children; Aetiology
Introduction
Although rotavirus is known to be the most common cause of diarrhoea in the under-five population, other viral, bacterial and protozoan enteric pathogens also contribute to the burden of diarrhoea. Following the World Health Organisation recommendation, several countries have introduced rotavirus vaccines (RVs) into their national immunisation schedules with an observed significant impact on reduction of rotavirus associated morbidity and mortality [1]. Zambia implemented RVs in the national expanded programme on immunisation starting in Lusaka province in 2012 and scaling up to country-wide by end of 2013; becoming the 17th GAVI eligible country to do so [2]. Over the period, significant reduction in acute gastroenteritis-associated in hospital morbidity and mortality has been recorded with the greatest reduction observed among children aged <1 year [3].
The increase in the number of countries introducing RV validates the fact that rotavirus is the leading cause of diarrhoea [4]; however, this reality will change as RVs make broad impacts. Moving forward, these enteric pathogens could become significant causes of diarrhoeal illness [5,6]. Therefore, comprehensive understanding of the aetiology of diarrhoea is critical for guiding control and therapeutic options, assessing vaccine performance and informing vaccine development priorities. For example, the value of individual or combination vaccines such as those in development against typhoid, ETEC and Shigella spp [7-11] would be better appreciated when there is data on the burden of disease. In this context, documenting the emerging top causes of diarrhoea post RV roll out should inform priorities for future diarrhoea disease prevention and control, and enteric vaccine development and implementation.
In this study, we aimed to document viral, bacterial, and protozoan enteric pathogens responsible of causing moderate-to-severe diarrhoea among children presenting at public health facilities in Zambia following the introduction of rotavirus vaccination.
Methods
Study design and participants
We used existing clinical data and raw stool samples collected for the purpose of evaluating the effectiveness of rotavirus vaccination under the “Programme for Awareness and Elimination of Diarrhoea (PAED)” in Zambia. The design of the original study was described in detail elsewhere [2]. Briefly, children were eligible if they were aged 0-59 months; admitted to the inpatient department or under care in the outpatient department; and caregiver verbal confirmation of child presenting with diarrhoea (defined as the passing of ≥3 abnormally loose stools in the past 24 hrs). Additionally, children were considered eligible if there was an indication of potentially severe diarrhoea by confirmation of at least 1 of the following symptoms assessed through physical examination by the study nurse or verbal confirmation by the caregiver: sunken eyes, loss of normal skin turgor, intravenous rehydration prescribed/administered, blood in stool and hospitalization for diarrhoea or dysentery. Enrolment of children was done during the early months post RV introduction between July 2012 and October 2013 in Lusaka province from health facilities. Detailed clinical, demographic, and vaccination data were systematically collected from each child including a raw stool sample collected at enrolment. Stool samples from children that remained after the RV effectiveness evaluation in Lusaka were stored at -80ºC. Our analysis sample included children from whom raw stool samples for enteric pathogen testing and complete clinical data were available.
Laboratory analysis
The qualitative, multiplex, polymerase chain reaction (PCR) based Luminex x-TAG® gastrointestinal pathogen panel (GPP) (Luminex Corporation, Austin TX, USA) was used to simultaneously detect 15 enteric pathogens i.e. viruses, bacteria and protozoa from the raw stool samples according to manufacturer’s protocol. Briefly, stool samples were first thawed and homogenized by bead beating in SK38 soil grinding tubes (Cat KT03961-1-006.2, Bertin Corporation, USA), incubated in NucliSENS® easyMAG® lysis buffer (BioMérieux, France) containing bacteriophage MS2 as an internal positive control and centrifuged. Resulting supernatant was then used for total nucleic acid extraction using the commercially available QIAamp MinElute Virus Spin Kit (Qiagen, Germany) and extracts were stored at -20ºC. For the simultaneous detection of enteric pathogens by GPP, 10µL of the stool nucleic acid extract was added as template to a 15µL x-TAG® GPP PCR reaction mix containing oligonucleotide primers targeting enteric pathogens: Adenovirus 40/41, Campylobacter (C. jejuni, C. coli and C. lari only), Clostridium difficile (C. difficile) toxin A/B, Cryptosporidium (C. parvum and C. hominis only), Entamoeba histolytica (E. histolytica), Escherichia coli (E. coli) O157, Enterotoxigenic E. coli (ETEC) LT/ST, Giardia (G. lamblia/G. intestinalis/G. duodenalis only), Norovirus GI/GII, Rotavirus A, Salmonella, Shiga-like Toxin producing E. coli (STEC) stx 1/stx 2, Shigella (S. boydii, S. sonnei, S. flexneri and S. dysenteriae), Vibrio cholerae (V. cholerae) and Yersinia enterocolitica (Y. enterocolitica). Three negative controls containing nuclease free water as template were included during each x-TAG® GPP PCR and the assay was run on a thermocycler (Applied Biosystems® 2720) under the following conditions: Initial denaturation at 95°C for 15 mins followed by 38 cycles of denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds and elongation at 72°C for 30 seconds with final extension at 72°C for 2 minutes and infinite hold at 4°C. A 5 µL volume of the resulting PCR product was hybridised in a 95 µL reaction mix containing magnetic beads (20 µL) and Streptavidin-R-Phycoerythrin (SAPE) conjugate in reporting solution (75 µL), incubated for 45 minutes at 45°C before fluorescence was detected on a MAGPIX® instrument. Median fluorescence intensity values were analysed by the x-TAG® data analysis software (TDAS) to determine presence or absence of enteric pathogens.
Post-hoc confidence interval estimation
We had a total of 1,135 children with both diarrhoea symptoms data and GPP test results. The prevalence of moderate-to-severe diarrhoea in this sample was 18.9%. To estimate the confidence interval, we used simple asymptomatic formula [12,13] based on the normal approximation to the binomial in PASS 14 (NCSS Inc.). Therefore, a sample size of 1,135 produces a two-sided 97% confidence interval with a width equal to 0.050 when the sample proportion is 0.189.
Outcome measurement
Moderate-to-severe diarrhoea: Using modified Vesikari score components [14], we defined moderate-to-severe diarrhoea as a score ≥ 9. The modified Vesikari score had seven components namely: diarrhoea duration, maximal number of diarrhoeal stools per 24 h period, vomiting duration, maximal number of vomiting episodes per 24 h period, maximal recorded fever, health care provider visits, and treatment. Each component or domain were rated on a scale of 0 to 3 points.
Statistical analysis
We summarised background characteristics using proportions or median as appropriate. We calculated the prevalence of enteric pathogens as the number of children with enteric pathogen infection divided by the total number of children under five included in the analysis. We also calculated the proportion of children with moderate-to-severe diarrhoea as the number of children with moderate-to-severe diarrhoea divided by the total number of children under five included in the analysis. 95% confidence intervals were estimated using normal approximation to binomial. We estimated pathogen-specific burden of moderate-to-severe diarrhoea using population attributable fraction [15-17]. The population attributable fraction was estimated using punaf user-written command in Stata [18]. Prior to using the punaf command, we fitted logistic regression with moderate-to-severe diarrhoea as outcome variable and pathogens presence as independent variables. All analyses were performed using Stata 15 MP (StataCorp, College Station, TX, USA).
Ethical approval
This study was reviewed and approved by the University of Zambia Biomedical Research Ethics Committee and the University of North Carolina at Chapel Hill Institutional Review Board. The Zambian Ministry of Health provided the authorisation to conduct the research. Caregivers of participants provided written informed consent prior to initiation of study procedures.
Results
Characteristics of children and prevalence of moderate-to-severe diarrhoea
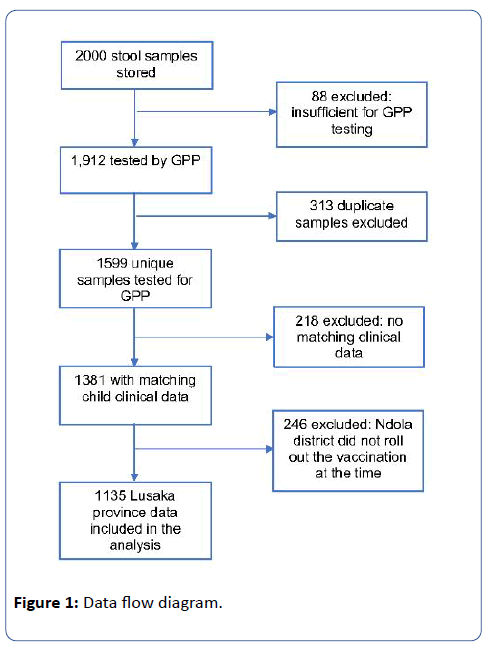
A total of 2,000 stool samples were stored from enrolled children of which 1,599 (80%) unique samples from individual children were available and sufficient for enteric pathogen testing by GPP. Matching clinical, demographic and vaccination data were available for 1,381 unique samples. Since RVs were not introduced in Ndola at the time the samples were collected, 246 samples from Ndola district were excluded from the analysis. The remaining 1,135 unique samples from Lusaka province were included in the final analysis (Figure 1). Of the 1,135 children included in our analysis, the median age was 14 months (IQR=8, 22), 59% lived in a home with inadequate WASH and 56% had received one or full dose rotavirus vaccination. (Table 1). The prevalence of moderate-t-severe diarrhoea was estimated as 18.9% (95%CI=16.7, 21.2) (Table 1). Higher proportion of infants (20.2%; 95%CI=16.9, 23.9) had moderate-to-severe diarrhoea compared to toddlers (19.1%; 95%CI=15.6, 23.2) and to children (15.2%; 95%CI=11.0, 20.5) (Table 1).
| Characteristics | Number of children (% of total) | No. (%) with moderate-to-severe diarrhoea | 95%CI |
|---|---|---|---|
| Age (months) | |||
| Median (IQR) | 14 (8, 22) | ||
| <12 (Infants) | 496 (44) | 100 (20.2) | (16.9, 23.9) |
| 12-23 (Toddlers) | 414 (36) | 79 (19.1) | (15.6, 23.2) |
| 24-59 (Children) | 224 (20) | 34 (15.2) | (11.0, 20.5) |
| Sex of Child | |||
| Female | 548 (48) | 112 (20.4) | (17.3, 24.0) |
| Male | 520 (46) | 92 (17.7) | (14.6, 21.2) |
| Vaccination Status | |||
| RV02 | 501 (44) | 96 (19.2) | (15.9, 22.9) |
| RV13 or Both | 633 (56) | 118 (18.6) | (15.8, 21.9) |
| WASH1 | |||
| Adequate | 471 (41) | 86 (18.3) | (15.0, 22.0) |
| Inadequate | 664 (59) | 128 (19.3) | (16.4, 22.5) |
| Total | 1135 (100) | 214 (18.9) | (16.7, 21.2) |
| 1Inadequate WASH was defined as households with unimproved source of drinking water as well as unimproved toilet facility [19]. 2RV0 means the child did not receive rotavirus vaccination; 3RV1 means the child received one dose of rotavirus vaccination. Both means the child received the full two doses of rotavirus vaccination. | |||
Table 1: Percentage of children with moderate-to-severe diarrhoea by background characteristics.
Prevalence of enteric pathogens and pathogen-specific burden of moderate-to-severe diarrhoea
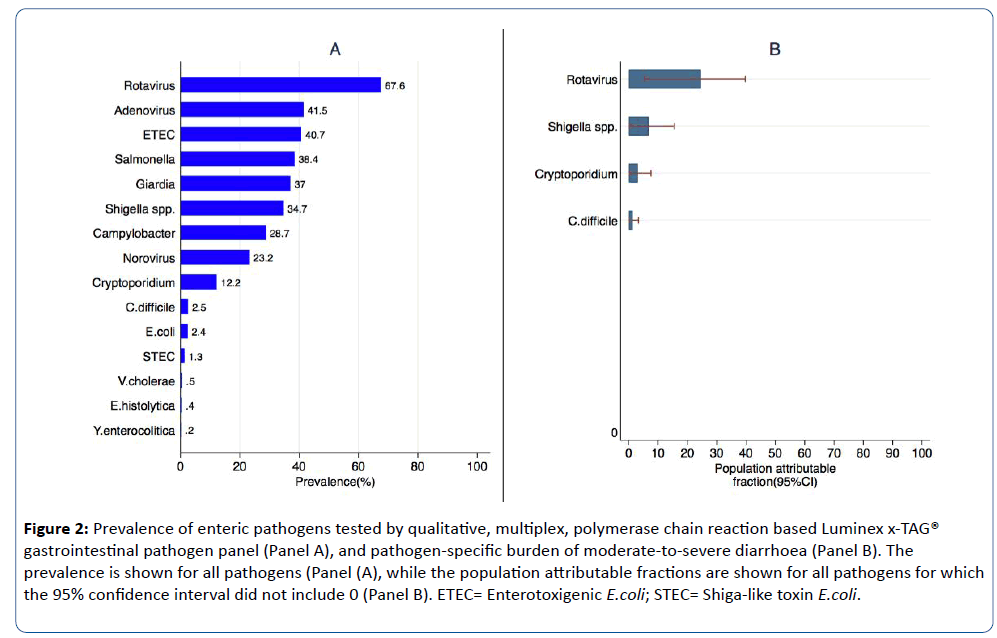
The top 5 enteric pathogens detected among children were rotavirus (67.6%), Adenovirus (41.5%), ETEC (40.7%), Salmonella (38.4%), and Giardia (37.0%) (Figure 2 Panel A). About 24.5% {95%CI=(5.4, 39.7)} of moderate-to-severe diarrhoea among children infected with rotavirus were attributable to rotavirus infection whereas about 6.7% {95%CI=(0.1, 15.5)} of moderate-to-severe diarrhoea among children infected with Shigella spp. were attributable to Shigella spp. (Figure 2 Panel B).
Figure 2: Prevalence of enteric pathogens tested by qualitative, multiplex, polymerase chain reaction based Luminex x-TAG® gastrointestinal pathogen panel (Panel A), and pathogen-specific burden of moderate-to-severe diarrhoea (Panel B). The prevalence is shown for all pathogens (Panel (A), while the population attributable fractions are shown for all pathogens for which the 95% confidence interval did not include 0 (Panel B). ETEC= Enterotoxigenic E.coli; STEC= Shiga-like toxin E.coli.
Discussion
We simultaneously identified important diarrhoeal aetiological pathogens (viral, bacterial and protozoan) in children under five in Zambia using a molecular based method. The five most common enteric pathogens detected among children were rotavirus (67.6%), Adenovirus (41.5%), ETEC (40.7%), Salmonella (38.4%), and Giardia (37.0%). The prevalence of moderate-to-severe diarrhoea in our sample was estimated as 18.9%. The two most attributable pathogens were rotavirus followed by Shigella where about 24.5% of moderate-to-severe diarrhoea among children infected with rotavirus were attributable to rotavirus infection and about 6.7% of moderate-to-severe diarrhoea among children infected with Shigella spp. were attributable to Shigella spp.
The importance of rotavirus was reaffirmed as the most high burden pathogen in our sample, which is consistent with what Kotloff and colleagues [16] found in the original GEMS study. This confirms that rotavirus remained the number one aetiology of moderate-to-severe diarrhoea requiring hospitalization in our setting despite RV introduction, which further highlights the importance of continuing efforts to monitor the effectiveness of vaccine introduction. While we have a recent paper from the university Teaching Hospital in Lusaka reporting sustained impact of RV on diarrhoeal hospitalization, the same paper reported substantial spikes in rotavirus positivity in stools by EIA in 2016 [20]. The more sensitive PCR detection methods would no doubt pick up greater frequencies of rotavirus infections if applied to the same samples. As we previously showed [21], comorbidity is common in our environment and indeed our results here further reveal the multiplicity of enteric pathogens potentially contributing to disease. The question of the clinical relevance of isolated pathogens in diarrhoeal stools thus still remains important warranting further interrogation.
Our data also hints at the possibility for combined or indeed shifting aetiologies post-RV, given the high burden of other enteric pathogens. The high prevalence of single Adenovirus infection, only next to rotavirus, in our study tallies with the GEMS re-analysis study where, when assessed with PCR-based methods, Adenovirus incidence was five times more than previously reported using microbiological methods [17,22], shifting this enteropathogen to become among the top six diarrhoea attributable pathogens [23,24]. However, our study did not find attributable cases of moderate-to-severe diarrhoea due to Adenovirus. Nonetheless, research is still required to understand the role of Adenovirus in moderate-to-severe diarrhoea in tropical setting as ours given its frequency. While the high burden of diarrhoea attributable to rotavirus and Shigella spp. as seen in our study may be due to the high sensitivity of the molecular based diagnosis method we used in this study, other reports such as Liu and colleagues [22] and many other studies [25-29] have noted that Shigella spp. was under detected in cultures compared to PCR.
Our study has important strengths worth noting. It is among the few studies that investigated a broad array of viral, bacterial, and protozoa enteric pathogens simultaneously using the PCR-based Luminex x-TAG GPP. Our study offers a more generalisable understanding of burden and aetiology of diarrhoea. We have reported diarrhoea aetiology in context of routine rotavirus vaccination.
We, however, recognised some limitations. First, the Luminex x-TAG GPP gives a qualitative readout which cannot account for the infectious load effect. This means our interpretation of the contribution to disease by the mere presence of a pathogen could be overestimated. Second, although this molecular assay detects multiple pathogens at once, species for some of the major ones are grouped together and contribution of each type to diarrhoea cannot be ascertained. (e.g. ETEC LT vs ST types, Norovirus GI/GII, or the Shigella. Third, our measure of diarrhoea included fever, which could have been induced by other comorbidities which are very common in our setting such as respiratory tract infections. Lastly Individuals can shed DNA when asymptomatic and for some time post clinical episode [30,31], which may not be attributed to the present episode of diarrhoea but detected by the molecular assay. Nonetheless, our effort to look at the aetiology of diarrhoea in our setting is a great achievement in documenting clinically relevant enteric pathogens.
For future studies on aetiology of diarrhoea, we propose that the quantitative PCR (qPCR) based assays are used to complement identification efforts by providing burden, species and strain information on enteric pathogens which can help with determining the attributable fraction each detected pathogen contributes to diarrhoea [22].
Conclusions
We found that about one-third of moderate-to-severe diarrhoea among children were attributable to rotavirus and shigella spp. The attribution of Shigella to diarrhoea observed in our population could help prioritise the development and process of introducing vaccines against Shigella in the post rotavirus vaccine era.
Financial Disclosure
The original case-control study was funded by the Bill & Melinda Gates Foundation (Grant # OPP1033211), and this aetiology evaluation study was funded by the Department for International Development (DFID) through Sanitation and Hygiene Applied Research for Equity (SHARE) Consortium at the London School of Hygiene and Tropical Medicine (Grant # ITDCHA2310/CD.9.08/SA.11.01).
Conflict of Interest
All authors declared that they have no conflict of interest.
Acknowledgment
We are grateful to all the study participants and their caregivers. We would like to thank Joyce Chilekwa and Annie Chikombo for their direct administrative efforts on this study. The authors also thank laboratory research intern Harriet Ng’ombe for her contribution towards the experimental work.
Data availability
Data will be made available to any interested researchers upon request. The CIDRZ Ethics and Compliance Committee is responsible for approving such request. To request data access, one must write to the Committee chair/Chief Scientific Officer, Dr. Roma Chilengi, (Roma.Chilengi@cidrz.org) or the Secretary to the Committee/Head of Research Operations, Ms. Hope Mwanyungwi (Hope.Mwanyungwi@cidrz.org) through the corresponding author (Samuel Bosomprah, PhD) mentioning the intended use for the data. The corresponding author will then facilitate express authorization to release the data as requested. Dataset request must include contact information, a research project title, and a description of the analysis being proposed as well as the format it is expected. The requested data should only be used for the purposes related to the original research or study. The CIDRZ Ethics and Compliance Committee will normally review all data requests within 48-72 hrs (Monday-Friday), and provide notification if access has been granted or additional project information is needed, before access can be granted.
References
- Parashar UD, Johnson H, Steele AD, Tate JE (2016) Health Impact of Rotavirus Vaccination in Developing Countries: Progress and Way Forward. Clin Infect Dis 62(suppl_2): S91-S5.
- Beres LK, Tate JE, Njobvu L, Chibwe B, Rudd C, et al. (2016) A Preliminary Assessment of Rotavirus Vaccine Effectiveness in Zambia. Clin Infect Dis 62(suppl_2): S175-S82.
- Mpabalwani EM, Simwaka CJ, Mwenda JM, Mubanga CP, Monze M, et al. (2016) Impact of Rotavirus Vaccination on Diarrheal Hospitalizations in Children Aged <5 Years in Lusaka, Zambia. Clin Infect Dis 62(Suppl 2): 183-187.
- ROTA Council, John Hopkins Bloomberg School of Public Helath, Center IVA. https://rotacouncil.org/vaccine-introduction/global-introduction-status/.
- Nitiema LW, Nordgren J, Ouermi D, Dianou D, Traore AS, et al. (2011) Burden of rotavirus and other enteropathogens among children with diarrhea in Burkina Faso. Int J Infect Dis 15: e646-e652.
- Becker-Dreps S, Bucardo F, Vilchez S, Zambrana LE, Liu L, et al. (2014) Etiology of childhood diarrhea after rotavirus vaccine introduction: a prospective, population-based study in Nicaragua. Pediatr Infect Dis J 33: 1156-1163.
- Feasey NA, Levine MM (2017) Typhoid vaccine development with a human challenge model. Lancet 390: 2419-2421.
- Riddle MS, Kaminski RW, Di Paolo C, Porter CK, Gutierrez RL, et al. (2016) Safety and Immunogenicity of a Candidate Bioconjugate Vaccine againstShigella flexneri 2a Administered to Healthy Adults: a Single-Blind, Randomized Phase I Study. Clin Vaccine Immunol 23: 908-917.
- Svennerholm AM, Tobias J (2008) Vaccines against enterotoxigenic Escherichia coli. Expert Rev Vaccines 7: 795-804.
- Taylor DN, Trofa AC, Sadoff J, Chu C, Bryla D, et al. (1993) Synthesis, characterization, and clinical evaluation of conjugate vaccines composed of the O-specific polysaccharides of Shigella dysenteriae type 1, Shigella flexneri type 2a, and Shigella sonnei (Plesiomonas shigelloides) bound to bacterial toxoids. Infect Immun 61: 3678-3687.
- Walker RI (2015) An assessment of enterotoxigenic Escherichia coli and Shigella vaccine candidates for infants and children. Vaccine 33: 954–965.
- Fleiss JL, Levin B, Paik MC (2003) Statistical Methods for Rates and Proportions. New York: John Wiley & Sons.
- Newcombe RG (1998) Two-Sided Confidence Intervals for the Single Proportion: Comparison of Seven Methods. Stat Med 17: 857-872.
- Freedman SB, Eltorky M, Gorelick M (2010) Pediatric Emergency Research Canada Gastroenteritis Study G. Evaluation of a gastroenteritis severity score for use in outpatient settings. Pediatrics 125: e1278-e1285.
- Blackwelder WC, Biswas K, Wu Y, Kotloff KL, Farag TH, et al. (2012) Statistical Methods in the Global Enteric Multicenter Study (GEMS). Clin Infect Dis 55: S246–S253.
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382: 209–222.
- Operario DJ, Platts-Mills JA, Nadan S, Page N, Seheri M, et al. (2017) Etiology of Severe Acute Watery Diarrhea in Children in the Global Rotavirus Surveillance Network Using Quantitative Polymerase Chain Reaction. J Infect Dis 216: 220–227.
- Newson RB (2013) Attributable and unattributable risks and fractions and other scenario comparisons. Stata J 13: 672-698.
- Central Statistical Office (CSO) (Zambia), Ministry of Health (MOH) (Zambia), ICF International (2014) Zambia Demographic and Health Survey 2013-14. Rockville, Maryland, USA.
- Mpabalwania EM, Simwaka JC, Mwenda JM, Matapo B, Parashar UD, et al. (2018) Sustained impact of rotavirus vaccine on rotavirus hospitalisations in Lusaka, Zambia, 2009–2016. Vaccine.
- Howard LM, Mwape I, Siwingwa M, Simuyandi M, Guffey MB, et al. (2017) Norovirus infections in young children in Lusaka Province, Zambia: clinical characteristics and molecular epidemiology. BMC Infect Dis 17: 92.
- Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, et al. (2016) Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388: 1291–1301.
- Adam MA, Wang J, Enan KA, Shen H, Wang H, et al. (2018) Molecular Survey of Viral and Bacterial Causes of Childhood Diarrhea in Khartoum State, Sudan. Front Microbiol 9: 112.
- Stockmann C, Pavia AT, Graham B, Vaughn M, Crisp R, et al. (2017) Detection of 23 Gastrointestinal Pathogens Among Children Who Present With Diarrhea. J Pediatric Infect Dis Soc 6: 231-238.
- von Seidlein L, Kim DR, Ali M, Lee H, Wang X, et al. (2006) A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Medicine 3: e353.
- Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, et al. (2015) Multucenter evaluation of the BioFire Filmarray Gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53: 915–925.
- Duong VT, Phat VV, Tuyen HT, Dung TT, Trung PD, et al. (2016) Evaluation of Luminex xTAG gastrointestinal pathogen panel assay for detection of multiple diarrheal pathogens in fecal samples in Vietnam. J Clin Microbiol 54: 1094–1100.
- Harrington SM, Buchan BW, Doern C, Fader R, Ferraro MJ, et al. (2015) Multicenter evaluation of the BD Max Enteric Bacterial Panel PCR assay for rapid detection of Salmonella spp., Shigella spp., Campylobacter spp., and Shiga Toxin 1 and 2 gens. J Clin Microbiol 53: 1639–1647.
- Lindsay B, Ochieng JB, Ikumapayi UN, Toure A, Ahmed D, et al. (2013) Quantitative PCR for detection of Shigella improves ascertainment of Shigella burden in children with moderate-to-severe diarrhoea in low-income countries. J Clin Microbiol 51: 1740–1746.
- Mukhopadhya I, Sarkar R, Menon VK, Babji S, Paul A, et al. (2013) Rotavirus Shedding in Symptomatic and Asymptomatic Children using Reverse Transcription-Quantitative PCR. J Med Microbiol 85: 1661-1668.
- Bines JE, Danchin M, Jackson P, Handley A, Watts E, et al. (2015) Safety and immunogenicity of RV3-BB human neonatal rotavirus vaccine administered at birth or in infancy: A randomised, double-blind, placebo-controlled trial. Lancet Infect Dis 15: 1389-1397.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences