Epigenetics Reprogramming of Autophagy is involved in Childhood Acute Lymphatic Leukemi
Dahb M Hassen, Khalid Bassiouny, Farha El-Shenawy, Hany Khalil
DOI10.21767/2573-0282.10045
Dahb M Hassen1, Khalid Bassiouny1, Farha El-Shenawy2 and Hany Khalil1*
1Department of Molecular Biology, Genetic Engineering and Biotechnology Research Institute, University of Sadat City, Egypt
2Department of Clinical Pathology, Faculty of Medicine, Mansura University, Egypt
- *Corresponding Author:
- Hany Khalil
Department of Molecular Biology
Genetic Engineering and Biotechnology Research Institute
University of Sadat City, Egypt
Tel: +20273291693
Email: hkhalil74@gmail.com
Received date: February 15, 2017; Accepted date: June 03, 2017; Published date: June 09, 2017
Citation: Hassen DM, Bassiouny K, El-Shenawy F, Khalil H (2017) Epigenetics Reprogramming of Autophagy is involved in Childhood Acute Lymphatic Leukemia. Pediatric Infect Dis 2:45. doi: 10.21767/2573-0282.100045
Copyright: © 2017 Hassen DM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Keywords
Acute lymphatic leukemia; DNA methylation; Autophagy
Introduction
Acute lymphoblastic leukemia (ALL) is one type of leukemia, or white blood cell cancer which normally characterized by excess lymphoblasts. Notably, in ALL, malignant immature white blood cells are continuously multiply and overproduced in the bone marrow. ALL causes damage and death by crowding abnormal cells in the bone marrow and spreading infiltrating to other organs. ALL is most common in childhood with a peak incidence at 2-5 years of age, and another peak in old age. The overall cure rate in children is about 80% in worldwide, and about 45%-60% of adults have long-term disease-free survival [1]. One the other hand, epigenetics is the study of heritable changes in cellular phenotype and gene expression caused by mechanism independent the original DNA sequences such as DNA methylation [2]. Several studies indicated the essential role of DNA methylation and epigenetic exchanges in various types of cell cancer. Interestingly, the Ras-association family (RASSF) of tumour suppressor genes have been found epigenetically inactivated in ALL via DNA over-methylation [3]. DNA methylation is an epigenetic mechanism that commonly used by cells to control gene expression. DNA methylation mainly occurs on cytosine at position 5 in a CpG dinucleotide which is mediated by DNA methyltransferases (DNMTs). Five forms of methyl-transferases exist; DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L. DNMTs work together either as de novo DNMTs, establishing the methyl group pattern on a sequence of DNA or as maintenance DNMTs that copy the methylation pattern on an existing strand of DNA to its new partner following replication. Methylation is sparse but global in mammals, found in CpG sequences across the entire genome, aside from certain stretches where the content of CpG is high (CpG islands) [4]. Methylation of CpG islands promoter suppresses the expression of corresponding genes due to poor recognition by transcription factors [4,5]. Consequently, the DNA methylation pattern modifies a certain protein expression profile which is ultimately associated with various human aging disorders such as Alzheimer’s (AD), and carcinogenicity [6]. Autophagy processes is characterized by accumulation of double-membraned cytoplasmic vacuoles regulating degradation events and recycling of cellular contents by delivering cytoplasmic materials, required for degradation, to lysosomes. Moreover, autophagy has been shown to play an important role in cell growth, development and disease pathology [7]. Notably, several studies indicated the essential role of autophagic machinery in regulating viral infection [8]. Activation of autophagy machinery is one of the down-stream events of a class III phosphatidylinositl I 3-kinase (PI 3-K) complex. The conjugations between more than 20 Atg proteins have been identified in autophagosome formation recruited from either endoplasmic reticulum (ER) or pre- autophagosomal structure (PAS). Atg5 and Atg12 are essential to initiate the autophagy vacuoles by recruiting other proteins to autophagosomal membrane from cytosol. The generation of phosphatidylinositol (3,4,5)- triphosphate (PIP-3,4,5) which recruits additional Atg proteins such as Atg8 (LC3) in conjugate system is essential for autophagosome elongation and maturation [9,10]. Finally, autophagosome fuses with lysosome containing hydrolytic enzymes that degrade the contents which are recycled for use in protein synthesis and energy production [11-14]. In this study we aim to investigate the possible involvement of epigenetic modification and DNA methylation of autophagy related molecules in peripheral blood samples that derived from children with ALL.
Materials and Methods
Samples conditions
Blood samples were collected from children with ALL in addition to control samples that were obtained from healthy individuals. The samples from ALL children were randomly collected from 57357 hospitals, Cairo, Egypt under full medical and ethical conditions. The blood samples were collected in ethylene diamine tetracetic acid (EDTA) and then were used to isolated genomic DNA and total RNA. Diagnostic of ALL connected parameters was performed by quantitative and qualitative ELISA of the most common enzymes such as terminal deoxynucleotidyl transferase, adenosine deaminase, 5'- nucleotidase, lactate dehydrogenase (LDH), purine nucleoside phosphorylase, acid phosphates, esterase, and hexosaminidase isoenzymes.
Gene amplification and resection enzymes digestion
Blood samples were collected in sodium citrate sterile tubes. Genomic DNA was extracted using G-spin Total Kit (Intron Biotechnology, Korea). Then the purified genomic DNA was used to amplify LC3B specific segment (500 nucleotides in length) using the specific primer sequences F: 5'- ACGCTGCGTGCCGCTGCTGGGTT-3' and R: 5'- TCTCACTCTCATACACCTCTGAG -3'. PCR was carried out in AmpGene DNA thermal cycler and the following PCR parameters were used, the initial denaturation step (95°C for 4 min), then 35 cycles (94°C for 40 sec, 60°C for 45 sec and 72°C for 60 sec). One unit of HpaII (Bio Basic Inc.) was used to digest 1 μg of PCR product of both ALL and control samples in total volume of 50 μl and one hour incubation at 37°C [15]. Finally, the digested product has been electrophoresed on 1% agarose gel for 30 min and monitored by gel documentation system.
Detection of methylation activity
Genomic DNA ALL derived samples was isolated using G-spin Total Kit (Intron Biotechnology, Korea). To investigate the possible methylated molecules of autophagy related genes Atg5 and LC3B, specific methylated primers for Atg5 and LC3B (Qiagen) were used to detect fold changes of methylated molecules in ALL samples compared with healthy individual samples. EpiTect Methyl II DNA Restriction Kit (Qiagen) was used to digest autophagy genes based on the methylation sensitive and dependent restriction sites. Fold changes of methylated molecules was calculated according the manufacture protocol [6].
Quantitative real time polymerase chain reaction analysis
Quantitative real time polymerase chain reaction (q-RT-PCR) is a sensitive method for the quantification of mRNA expression levels using specific primers. Q-RT-PCR will be used to detect the expression levels of autophagy related genes as one of mentoring methods to investigate the formation of autophagosome in ALL derived samples. RNA is first reverse transcribed into complementary DNA (cDNAWQ) using reverse transcriptase. The resulting cDNA is used as template for subsequent PCR amplification using DNA polymerase enzyme and primers specific for one or more genes. The relative expression of DNMT1, DNMT2, DNMT3, Atg5, and LC3 B were detected using the QuantiTect SYBR Green PCR Kit (Qiagen) and oligonucleotides specific for each individual gene (Table 1). Levels of GAPDH were amplified using specific oligonucleotides, GAPDH-For-5'-TGGCATTGTGGAAGGGCTCA-3' and GAPDH-Rev-5'- TGGATGCAGGGATGATGTTCT-3' which was used for normalization.
| Description | Primer sequences 5'-3' |
|---|---|
| DNMT1-For | CCCATGCATAGGTTCACTTCCTTC |
| DNMT1-Rev | TGGCTTCGTCGTAACTCTCTACCT |
| DNMT2-For | CATACAATGCCCGTGTGAGTTCTTAAGG |
| DNMT2-Rev | CGTGTGTCTAAATGGCTTGAGTACAGT |
| DNMT3-For | TGCAATGACCTCTCCATTGTCAAC |
| DNMT3-Rev | GGTAGAACTCAAAGAAGAGGCGG |
| Atg5-For | CGTGTATGAAAGAAGCTGATGC |
| Atg5-Rev | ACGAAATCCATTTTCTTCTGGA |
| LC3B-For | CGTCCTGGACAAGACCAAGT |
| LC3B-Rev | CCATTCACCAGGAGGAAGAA |
Table 1: Oligonucleotide sequences used for detection of indicated genes at RNA level.
The following parameters have been used in RT-PCR program, 95°C for 5 min, 35 cycles (94°C for 45 sec, 60°C for 30 sec and 72°C for 30 sec) and finally 72°C for 10 minutes. The results were analyzed using ΔΔ Ct equations [6].
Statistical analysis
Microsoft Excel was used for statistical calculation, graphs, and histograms. SDS 2.2.2 software was used to analyze the qRTPCT data to drive the ΔΔCt values using the following equations; (Delta Ct)=Ct-sample–Ct-control. (ΔΔCt)=ΔCt–ΔCt- normalized control. Finally, the relative gene expression is equal 2-ΔΔct of the final values [6,9]. The following website has been used For HpaII digestion protocol https://www.neb.com/products/R0171-HpaII. For Atg5 gene sequences and analysis the following website has been used; https://www.ncbi.nlm.nih.gov/pubmed.
Results
Resting Blood samples derived from children with ALL exhibit low expression of autophagy related genes
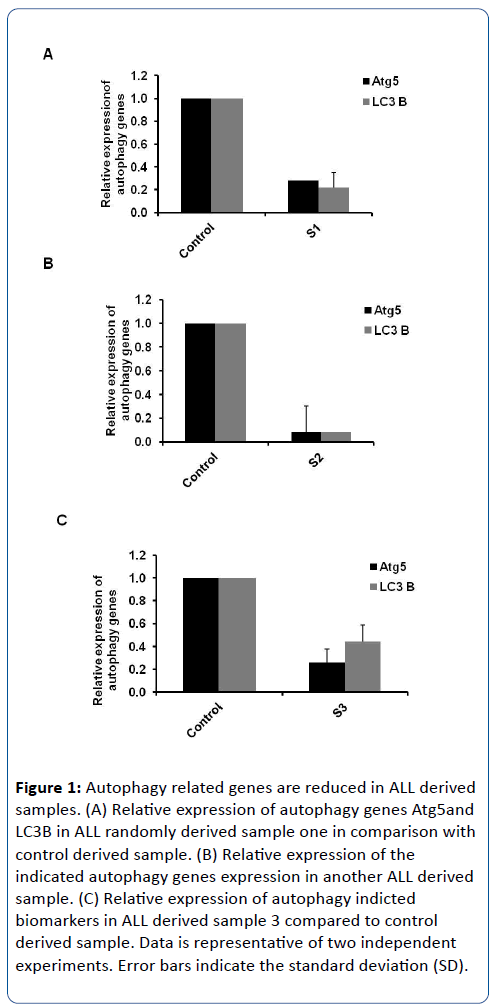
To investigate the kinetics of autophagosome formation in response to ALL, three different samples that randomly obtained from children with ALL were investigated for the relative expression of autophagy Atg5 and LC3B genes. The conjugation between Atg5 and Atg12 is essential for autophagy initiation step in cooperation with autophagy Atg16 protein. The interruption of Atg5 expression profile is sufficient to inhibit the autophagosome formation [6,16]. During elongation step, the conversion of cytosolic isoform of LC3 protein (autophagy Atg8 protein) to membraned isoform of LC3 is required to complete the formation of mature autophagosomes. Increasing of LC3B at RNA or protein levels can be used to prove the induction of autophagy [17]. Accordingly, total RNA and complementary cDNA have been performed from ALL randomly obtained samples in addition to healthy obtained samples. The relative expression of autophagy Atg5 at RNA level was detected in ALL samples using specific oligonucleotides to figure out the possible initiation of autophagosome formation during ALL disorder. Additionally, the relative expression of LC3B was detected by using specific oligonucleotides to check the status of autophagosome elongation process. Interestingly, the relative expression of both autophagy related genes Atg5 and LC3 were strongly reduced in ALL obtained samples in comparison with obtained samples from healthy individuals (Figure 1A-C). This result indicates that the kinetics of autophagosome formation is interrupted in ALL derived samples and raises the inquiry for the possible biological reasons for this interruption.
Figure 1: Autophagy related genes are reduced in ALL derived samples. (A) Relative expression of autophagy genes Atg5and LC3B in ALL randomly derived sample one in comparison with control derived sample. (B) Relative expression of the indicated autophagy genes expression in another ALL derived sample. (C) Relative expression of autophagy indicted biomarkers in ALL derived sample 3 compared to control derived sample. Data is representative of two independent experiments. Error bars indicate the standard deviation (SD).
The expression level of DNMT1 is highly increased in ALL samples
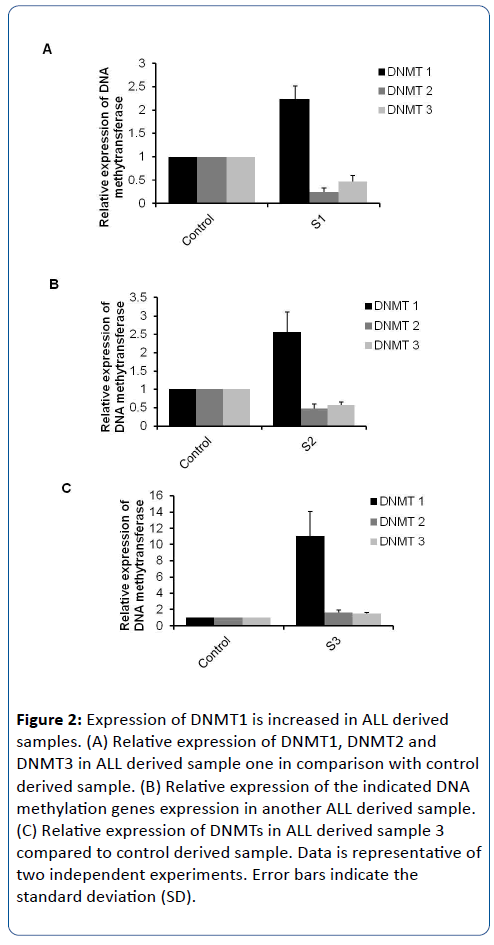
DNA methyltransferases (DNMTs), are responsible for transferring a methyl group from universal donor, S-adenosyl-Lmethionine (SAM), to the 5-position of cytosine remains in DNA molecules. Methylation activity of DNMTs plays critical role on various human diseases and medical disorders such as Alzheimer (AD) and carcinogenicity. Five forms of DNMTs have been identified; DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L [4,18]. Importantly, possible methylation of CpG islands promoter inhibits the expression of corresponding genes due to poor recognition sites by transcription factors. Therefore, the specific oligonucleotides that targeted DNMT1 and DNMT2 in addition to oligonucleotides that targeted conserved sequences of different DNMT3 isoforms have been used to detect the relative expression of indicated DNMTs in ALL derived samples [6]. Interestingly, the relative expression of DNMT1 is increased in ALL selected samples in comparison with control samples while other DNMTs (DNMT2 and DNMT3) showed negligible differentiations (Figure 2A-C).
Figure 2: Expression of DNMT1 is increased in ALL derived samples. (A) Relative expression of DNMT1, DNMT2 and DNMT3 in ALL derived sample one in comparison with control derived sample. (B) Relative expression of the indicated DNA methylation genes expression in another ALL derived sample. (C) Relative expression of DNMTs in ALL derived sample 3 compared to control derived sample. Data is representative of two independent experiments. Error bars indicate the standard deviation (SD).
Moreover, in ALL sample-1 (S1), the expression level of DNMT1 was increased 2 fold compared to control derived sample. ALL Sample-2 (S2) showed 2.5 fold up regulation of DNMT1 compared with its expression on the control sample while the relative expression of DNMT1 in ALL sample-3 (S3) has been increased 10 fold in comparison with the healthy control sample (Figure 1A-C). Taken together, this result demonstrates that the expression profile of DNMT1 is increased in ALL derived samples and indicates the possible involvement of DNA methylation and epigenetics reprogramming of cellular genes on children with ALL.
Methylation of CpG in LC3B and Ag5 is recognized in promoter region and coding sequence
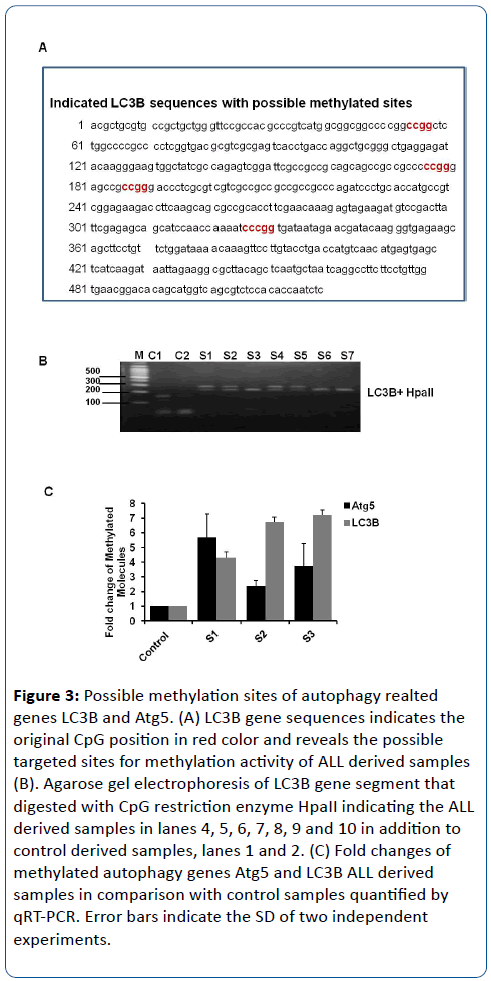
To confirm the possible epigenetic reprogramming of autophagy LC3B gene, HpaII restriction enzyme has been used to digest the indicated segment of LC3B gene. HpaII has been shown to cleave the sequence CCGG. Importantly, the cleavage with this restriction enzyme is blocked when the substrate DNA is methylated by CpG methylase [15]. Noteworthy, the sequence analysis of LC3B showed 4 different restriction sites that can be targeted by HpaII (Figure 3A). Therefore, genomic DNA has been isolated and purified from 10 samples that were derived from children with ALL in addition to 10 samples that were obtained from healthy individuals. The indicated segment of LC3B was amplified by using specific oligonucleotides and then was digested by using HpaII restriction enzyme. Interestingly, the digested ALL derived samples have been changed in size and number of indicated fragments in comparison with control samples. Moreover, the digested products of control samples showed three different fragments with molecular size of approximately 180, 170 and 75 nucleotides (nt) while ALL digested samples showed only two fragment with molecular size of approximately 220nt and 230nt (S1, S2, S4 and S6) or 250nt and 75nt (S3, S6 and S7) (Figure 2B). These data indicate that the digestion activity of HapII is interrupted due to methylation changes of cytosine nucleotides in its specific restriction sites and confirm the epigenetic reprogramming of Atg5 gene in ALL derived samples. Furthermore, we examined the methylated promoter region in autophagy related genes Atg5 and LC3B, and we found that Atg5 and LC3B promoters were methylated in ALL derived samples (Figure 3A-C). Collectively, our data here provide an evidence for methylation of essential autophagy genes LC3B and Atg5 in ALL samples and indicate that epigenetic reprogramming of autophagy genes is involved in ALL disease.
Figure 3: Possible methylation sites of autophagy realted genes LC3B and Atg5. (A) LC3B gene sequences indicates the original CpG position in red color and reveals the possible targeted sites for methylation activity of ALL derived samples (B). Agarose gel electrophoresis of LC3B gene segment that digested with CpG restriction enzyme HpaII indicating the ALL derived samples in lanes 4, 5, 6, 7, 8, 9 and 10 in addition to control derived samples, lanes 1 and 2. (C) Fold changes of methylated autophagy genes Atg5 and LC3B ALL derived samples in comparison with control samples quantified by qRT-PCR. Error bars indicate the SD of two independent experiments.
Discussion
In the current work we aim to investigate the possible involvement of epigenetics modification of autophagy in children with ALL. Therefore, blood samples were obtained from children with ALL and relative gene expression of DNMTs and autophagy related genes Atg5 and LC3B were detected at RNA using specific oligonucleotides. Our results showed low expression level of Atg5 and LC3B accompanied with increasing expression level of DNMT1 in ALL derived samples compared to healthy derived samples. Furthermore, digestion of the indicated sequences of LC3B gene segment with HpaII that targeted CCGG position showed partial activation of HpaII on ALL derived samples in comparison with control derived samples. These findings indicate the possible involvement of epigenetic reprogramming of autophagy related genes LC3B and Atg5 via activation of DNMT1. Additionally, here we provide easy and direct methods to confirm the methyaltion activity of specific DNA molecules by using HpaII restriction enzyme that targeted the CpG position and interrupted upon methyl alternation of cytosine nucleotides. DNA methylation is an epigenetic event that affects cell function by altering gene expression and refers to the covalent addition of a methyl group, catalyzed by DNA methyltransferase (DNMTs), to the 5- carbon of cytosine in a CpG dinucleotide. Methods for DNA methylation analysis can be divided roughly into two types: global and genespecific methylation analysis. For global methylation analysis, there are methods which measure the overall level of methyl cytosine in genome such as chromatographic methods and methyl accepting capacity assay. For gene- specific methylation analysis, a large number of techniques have been developed. Most early studies used methylation sensitive restriction enzymes to digest DNA followed by Southern detection or PCR amplification. Recently, bisulfite reaction based methods have become very popular such as methylation specific PCR (MSP), bisulfite genomic sequencing PCR [15,18]. Additionally, in order to identify unknown methylation hot-spots or methylated CpG islands in the genome, several of genome- wide screen methods have been invented such as Restriction Landmark Genomic Scanning for Methylation (RLGS-M), and CpG island microarray. Several studies have implicated the biological role and molecular interaction of DNMTs in cancer, aging and hepatitis C virus infection [8]. The expression of DNMT1 is increased in cancer and induces DNA methylation of various tumor suppressor genes. Beside some DNA methylation activities, DNMT1 catalyzes cytosine converts to thymine by adding methyl group in CG sequence. While, the expression of DNMT2 has been detected in oocytes, embryos, spermatocytes, and spermatids. Up-regulation of DNMT2 was reported to activate viability and fertility Additionally, DNMT2-modulated cellular signaling pathways have been demonstrated in health and human disease conditions [6]. Here, we found that DNMT1 is highly expressed in blood samples that derived from patients with ALL in association with methylation of the promoter region of Atg5 and LC3B leading to low expression level of indicated genes. Subsequently, our findings here demonstrate that initiation and elongation steps of autophagic machinery are interrupted in ALL samples that might be facilitate evaluation of this medical disorder. Noteworthy, autophagic induction is quantifiable by following microtubule-associated protein light-chain 3A or B (LC3A or LC3B) changes. The difference between LC3 A and B still not completely understood. Host cell LC3 is present in two forms, the cytosolic type LC3-I with an electrophoretic mobility corresponding to the relative molecular mass of 18 KDa, and membrane-associated form, LC3-II, with an apparent mass of 16 KDa. The latter form, LC3-II, inserts into the membrane of nascent autophagosomes and correlates with the appearance of LC3-positive autophagosomes [10,17]. The whole gene sequences of LC3B is approximately two thousand nucleotides with four different position of CCGG that are located in the first five hundred nucleotides. Accordingly, here we amplified the segment of LC3B with the four restriction sites of HpaII to investigate the digestion activities in ALL derived samples compared to control samples. Expectedly, the methylation activity of CpG positions in LC3B segment has been resulted in inactive digestion of HpaII indicating the possible epigenetic reprogramming of autophagy related genes in ALL disease.
References
- Rubnitz JE (2000) Molecular diagnostics in the treatment of childhood acute lymphoblastic leukemia. J Biol Regul Homeost Agents 14: 182-186.
- Dunwell TL, Hesson LB, Pavlova T, Zabarovska V, Kashuba V, et al. (2009) Epigenetic analysis of childhood acute lymphoblastic leukemia. Epigenetics 4: 185-193.
- Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, et al. (2009) The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer 8: 42.
- Robertson K D, Jones P A (2000) DNA methylation: past, present and future directions. Carcinogenesis 21: 461-467.
- Robertson KD, Wolffe AP (2000) DNA methylation in health and disease. Nat Rev Genet 1: 11-19.
- Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, et al. (2016). Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics 11: 381-388.
- Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self-eating. Cell Death and Differentiation 12: 1542-1552.
- Khalil H (2017) The intracellular signaling that associated with Influenza A virus infection. Pediatric Infectious Diseases 2: 38.
- Khalil H (2012) Influenza A virus stimulates autophagy to undermine host cell IFN-β production. Egyptian Journal of Biochemistry & Molecular Biology 30: 283-299
- Tazi M F, Dakhlallah DA, Caution K, Gerber MM, Chang SW, et al. (2016) Elevated Mirc1/Mir17-92 cluster expression negatively regulates autophagy and CFTR (cystic fibrosis transmembrane conductance regulator) function in CF macrophages. Autophagy 12: 2026-2037.
- Orvedahl A, Levine B (2008) Autophagy and viral neurovirulence. Cellular Microbiology 10: 1747-1756.
- Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ (2007) Autophagy and vacuole homeostasis - A case for self-degradation? Autophagy 3: 417-421.
- Yen WL, Klionsky DJ (2007) Atg27 is a second transmembrane cycling protein. Autophagy 3: 254-256
- Legakis JE, YenWL, Klionsky DJ (2007) A cycling protein complex required for selective autophagy. Autophagy 3: 422-432.
- Al-Younes HM, Al-Zeer MA, Khalil H, Gussmann J, Karlas A, et al. (2011) Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy 7: 814-828.
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T (2001) Beclin-phosphatidylinositol 3- kinase complex functions at the trans-Golgi network. Embo Reports 2: 330-335.
- Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA (2000b) Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res 28: 2108-2113.
- Robertson K D, Jones P A (2000) DNA methylation: past, present and future directions. Carcinogenesis 21: 461-467.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences